Global Journal of Cancer Therapy
A first comprehensive look at the order-disorder nature of RTK KIT native and carcinogenic targets
Luba Tchertanov* and Julie Ledoux
Cite this as
Tchertanov L, Ledoux J (2022) A first comprehensive look at the order-disorder nature of RTK KIT native and carcinogenic targets. Glob J Cancer Ther 8(1): 036-039. DOI: 10.17352/2581-5407.000046Copyright License
© 2022 Tchertanov L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Introduction
Receptors Tyrosine Kinases (RTKs) act as sensors for extracellular ligands, the binding of which triggers dimerization, activation, and autophosphorylation of specific tyrosine (Y) residues in the Cytoplasmic Domain (CD). This leads to the recruitment and activation of multiple downstream signaling proteins, which regulate various aspects of cellular physiology.
The physiological actions of RTK KIT, control cell survival, proliferation, differentiation, and migration, depending on the activation of specific or overlapping signaling pathways, which endows the activity of the SCF/KIT system (where SCF is the stem cell factor) with great complexity. The native KIT activation regulated by SCF triggers the initiation of multiple signal transduction pathways leading to normal transduction and transcription processes in the cells. Aberrant regulation of KIT signaling networks is associated with the progression of many cancer types, including human acute myeloid leukemia, aggressive systemic mastocytosis, melanoma, gastrointestinal stromal tumor, and stomach cancers. Disclosure of the KIT-activated pathways in carcinogenesis will be crucial in developing KIT-targeted therapies.
RTK KIT consists of an extracellular ligand-binding domain formed by five Ig-like fragments connected to a Cytoplasmic Domain (CD) by a transmembrane helix. RTK KIT regulates various crucial cellular processes via its modular CD composed of the Tyrosine Kinase Domain (TKD) that also has a modular architecture consisting of proximal (N-) and distal (C-) lobes linked by a Kinase Insert Domain (KID). The catalytic mechanism involves fragments from the TKD that regulate RTK activity – the Catalytic (C-) and the Activation (A-) loops, the Juxtamembrane Region (JMR), and the Cα-helix. At the same time, multiple phosphorylation sites associated with JMR, KID and C-terminal are the platforms of post-transduction processes, phosphorylation, intracellular protein binding, and downstream signaling.
Studying the transduction processes in RTK KIT using experimental techniques is complex and/or highly restricted. A theoretical description at the structural level is also limited or impossible because no experimental data describing a full-length KIT falls short of revealing the structural details of KID, JMR and C-tail at the atomistic level.
Our research focuses on (i) generation of the 3D model of the full-length CD of KIT; (ii) description of its structural and dynamical features and their relationships; (iii) delivery of targets helpful for alternative strategies to modulate KIT activation and signaling in native and mutated states. The proposed strategy targets KIT allosteric intra-molecular pockets and interfaces exposed to protein-protein interactions. Such an approach encouraged the development of highly specific pharmaceuticals to avoid side effects resulting from inhibition with non-specific ATP-competitive molecules (e.g. imatinib).
Methods
We performed in silico studies using different classical and state-of-the-art computation techniques. To build 3D models of the full-length KIT cytoplasmic domain in inactive, active states for the native KIT, and its constitutively active (D816V) mutant, we used a hybrid protocol that combines de novo techniques (Rosetta and MODELLER) and empirical crystallographic data (PDB: 1T45 and 1PKG). All generated models were studied by multiple extended (2-6 µs) Molecular Dynamics (MD) simulations (AMBER) under conditions that mimic KIT’s natural environment. Produced simulation big data were analyzed using canonical (conventional) and advanced innovative statistical methods to describe the structurally and dynamically related parameters, interpreted in terms of structure-dynamics-function relationships, Gibbs Free Energy, communication pathways, and allosteric regulation.
Results
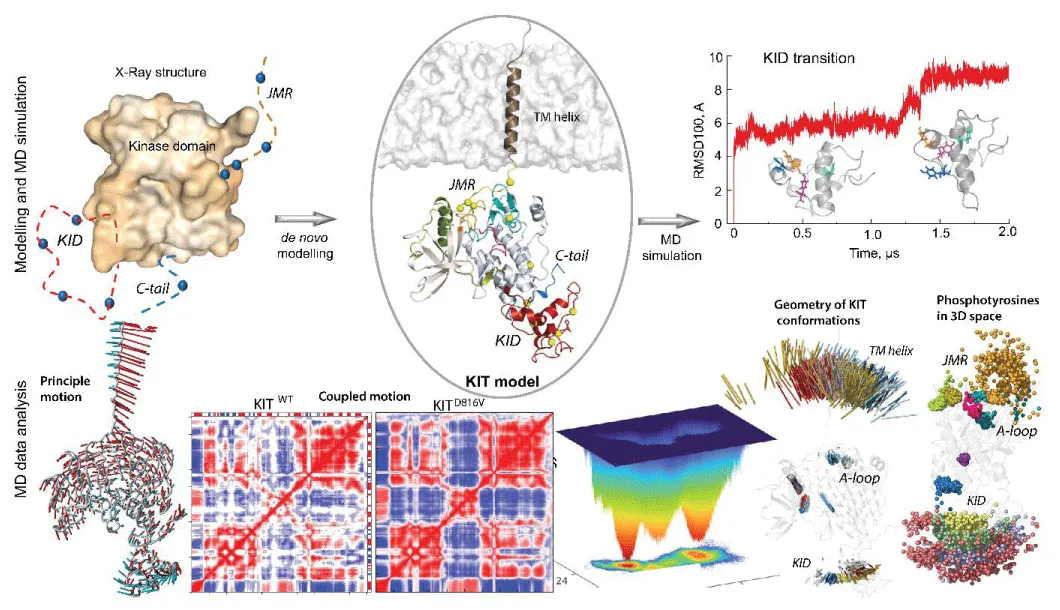
Our 3D models of a full-length CD of RTK KIT attached to a Tran’s Membrane (TM) helix (Figure 1) are the first description of KIT at the atomistic level [1,2]. This modeling is a cornerstone for characterizing KIT’s inherent properties (KIT DYNASOME) and interactions of the native KIT and oncogenic mutant D816V with signaling proteins (KIT INTERACTOME).
Accomplishing the accurate description of the multidomain KIT inherent characteristics at the atomistic level, we explained the role of somatic mutation D816V on the structural and dynamical properties of KIT [2,3]. We focused on its intrinsic (intra-domain) and extrinsic (inter-domain) disorder and their relationships. Conformational ensembles of native and mutated KIT were represented through free energy landscapes defining KIT’s most stable and intermediate conformations. We proved that the multidomain KIT is a modular protein consisting of a quasi-stable TKD crowned by at least four Intrinsically Disordered (ID) regions JMR, KID, A-loop and C-tail. Each ID domain structure represents a complex mixture of a wide variety of differently folded conformations describing a reversible folding-unfolding process specific for each ID domain. In particular, the KID, composed of transient helices linked by coils, is a KIT disordered domain but shows a globule-like shape stabilized by multiple non-covalent interactions (hydrogen bonding and van der Waals forces). Furthermore, KID displays different positions derived from two types of motions - linear (translation) and angular (rotation) displacement - regarding the stable TK domain. The elongated regions (JMR, A-loop, and C-tail) show local disorder, as evidenced by the alternating positions of their short segments relative to the stable TKD.
The two-level disorder (intrinsic and extrinsic) reflected on structural and dynamical properties provides high conformational variability of KIT and supplies the excellent adaptability of JMR, KID and C-tail for the scaffolding (docking sites) and recruitment of KIT partners (adaptors, signaling and scaffolding proteins). Consequently, the overall structure of KIT represents a continuous spectrum of conformations with different degrees and depths of the disorder, thereby generating a convoluted protein structural space. We also proved that the level of disorder of one ID domain depends (cross-correlations, highly coupled motions) on the other remote ID regions confirming their functional dependence, classified as allosteric regulation, a phenomenon widely observed in many proteins, particularly RTKs.
Comparing the KIT D816V mutant to the native KIT, we found that the KIT mutant shows a significantly increasing coupling of intra- and inter-domain motions. Moreover, the KIT communication pathways network is strongly different in the two proteins. Such multipanel characterization explained more explicitly the role of each region in maintaining the inactive, active, and constitutively active states. We established the relationships between phosphotyrosines containing ID regions, which are signaling carriers. We also suggested that the KIT inactive state encodes all properties of the active protein for post-transduction events. Such a hypothesis echoes the high dependence between INTERACTOME and DYNASOME of RTK KIT.
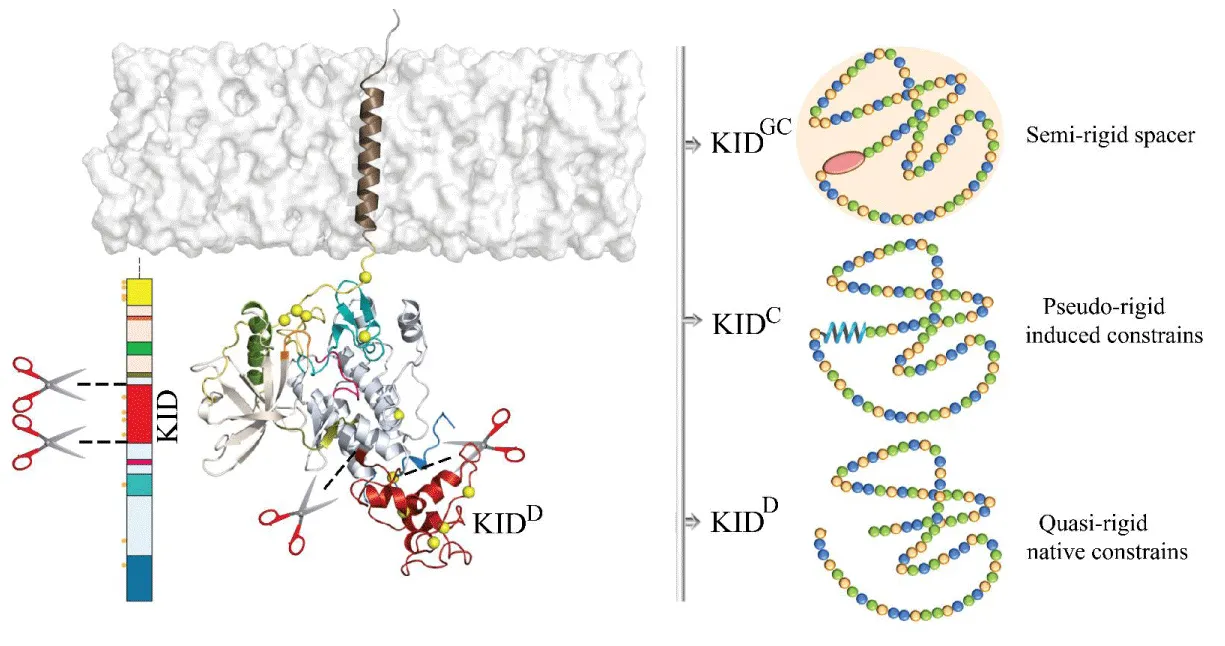
Next, we focused on KIT’s kinase Insert Domain (KID), the critical recruitment platform for a host of downstream signaling proteins-enzymes, adapters, and scaffolding proteins. We showed that the KIT KID is intrinsically disordered, and this property is disconnected from the KIT context, represented as a cleaved polypeptide or as a domain of the receptor [4]. The intrinsic disorder is a central feature in the function of numerous proteins, enabling them to serve as molecular switches and hubs in biological networks.
In all studied entities, KID folding displays a collection of highly variable helices altered in length and type of their secondary structures (α- or 310-helices or β-strands). Except for the long-lived αH1, KID helices are transient, converting between the α- and 310-helices or a helix and strand or non-regular secondary structures — turns, bends, and coils. Structural consistency of αH1-helix suggested that KID inner ordered motif is critical for the KIT function. Indeed, despite the changes in the αH1-helix length along the same trajectory or in different trajectories of distinct KID entities, αH1-helix, like a drop of glue, attaches the KID structural fragments around them by manifold contacts and stabilizes a ‘globule-like’ shape of the intrinsically disordered KID. The ‘organizing role’ of αH1-helix in stabilizing the KID structure was previously attributed to tyrosine Y747 located on the H4-helix [2]. We suggest that the Y747 and αH1-helix functions are complementary and can be mutually dependent.
To deliver the valid usable species as an initial template for empirical studies, a generic cyclic KID (KIDGC) of RTK KIT was proposed as an entity that could be best suited for future studies on the KIT post-transduction effects involving KID [5]. KIDGC is composed of the 80 amino acids cleaved polypeptide (F689–D768) cyclized by the insertion of four glycine residues acting as a physical connector or spacer between its N- and C-KID termini (Figure 2). We suggested that this generic cyclic KID is also an Intrinsically Disordered Protein (IDP). As the characterization of IDPs is not a trivial problem, we report the compiled structural description of KID and resort to the recapitulation of the available KIT KID data obtained for the different KID species to compare them in terms of Gibbs free energy. We assumed that these KID species have similar structural and dynamic characteristics indicating the intrinsically disordered nature of this domain. This finding means that both polypeptides, cyclic (KIDGC) and linear (KIDC), are valid models of KID integrated into the RTK KIT and will be helpful for further computational and empirical studies of post-transduction KIT events.
However, since the kinase domain is a central hub of both receptor activation and communication between distant functional regions-JMR, A-loop, KID, and C-terminal−, investigation of signal transduction mechanisms or the mechanisms of allosteric regulation of KIT in the native or mutated state would require full-length KIT or at least, its full-length cytoplasmic domain.
Finally, focusing on KID, we studied the phosphorylation effects on its properties [6]. We are now generating a molecular complex KIT-p85α N-terminal SH2 domain as the first precursor for studying KIT signal transduction mediated by KID. We plan to validate our predictions by comparing the de novo model with valuable empirical data reporting the complex of p85α PI3K regulatory subunit N-terminal SH2 domain with a KID peptide (PDB ID: 2IUH).
Our in silico-generated models of the allosterically regulated KIT in two native forms (inactive and active) and constitutively active state is the first step for reconstruction of its complete INTERACTOME, composed of a set of KIT complexes with its signaling proteins, and DYNASOME, constituting of an ensemble of KIT intermediate conformations before, over and after post-transduction processes upon its physiological and pathological context.
Our data will be used to search for alternative strategies for modulating KIT activation and signaling, particularly by targeting KIT allosteric intra-molecular pockets and interfaces in KIT-protein macromolecular complexes to avoid or decrease side effects resulting from inhibition with non-specific ATP-competitive molecules (e.g. imatinib).
- Inizan F, Hanna M, Stolyarchuk M, Chauvot de Beauchêne I, Tchertanov L. The First 3D Model of the Full-Length KIT Cytoplasmic Domain Reveals a New Look for an Old Receptor. Sci Rep. 2020 Mar 25;10(1):5401. doi: 10.1038/s41598-020-62460-7. PMID: 32214210; PMCID: PMC7096506.
- Ledoux J, Trouvé A, Tchertanov L. The Inherent Coupling of Intrinsically Disordered Regions in the Multidomain Receptor Tyrosine Kinase KIT. Int J Mol Sci. 2022 Jan 29;23(3):1589. doi: 10.3390/ijms23031589. PMID: 35163518; PMCID: PMC8835827.
- Ledoux J, Trouvé A, Tchertanov L. Does cancerogenic mutation D816V influence the intrinsic disorder of the modular receptor tyrosine kinase KIT? In preparation. 2023.
- Ledoux J, Trouvé A, Tchertanov L. Folding and Intrinsic Disorder of the Receptor Tyrosine Kinase KIT Insert Domain Seen by Conventional Molecular Dynamics Simulations. Int J Mol Sci. 2021 Jul 9;22(14):7375. doi: 10.3390/ijms22147375. PMID: 34298994; PMCID: PMC8307779.
- Ledoux J, Tchertanov L. Does generic cyclic Kinase Insert Domain of Receptor Tyrosine Kinase KIT clone its native homologue? Submitted as an invited paper. Int J Mol Sci. 2022.
- Ledoux J, Tchertanov L. How phosphorylation of tyrosine residues affects the local and global geometry of the receptor tyrosine kinase KIT. 2023.

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
