International Journal of Immunotherapy and Cancer Research
Regulation of Protein Degradation and Homeostasis by the Cytokine-Inducible Deubiquitinating Enzymes
Kwang-Hyun Baek*
Cite this as
Baek KH (2018) Regulation of Protein Degradation and Homeostasis by the Cytokine-Inducible Deubiquitinating Enzymes. Int J Immunother Cancer Res 4(1): 001-003. DOI: 10.17352/2455-8591.000019Ubiquitin-proteasome system (UPS) is the major signaling pathway responsible for regulating protein turnover in cells. Deubiquitinating enzymes (DUBs) have an important role in this signaling pathway by eliminating ubiquitins from substrates and inhibiting proteasomal degradation to maintain cellular homeostasis.
Based on the published data on cytokine-inducible DUBs including DUB-1, DUB-2, DUB-2A, DUB-1A and DUB-3, they play a pivotal role in the regulation of proliferative and apoptotic processes for immune cells.
In this review, I discuss the importance of cytokine-inducible DUBs and the development of new therapeutic targets for these DUBs in immune-related diseases.
Ubiquitin-proteasome system (UPS) is the major signaling pathway responsible for regulating protein turnover in cells. Deubiquitinating enzymes (DUBs) have an important role in this signaling pathway by eliminating ubiquitins from substrates and inhibiting proteasomal degradation to maintain cellular homeostasis.
Based on the published data on cytokine-inducible DUBs including DUB-1, DUB-2, DUB-2A, DUB-1A and DUB-3, they play a pivotal role in the regulation of proliferative and apoptotic processes for immune cells.
In this review, I discuss the importance of cytokine-inducible DUBs and the development of new therapeutic targets for these DUBs in immune-related diseases.
Introduction
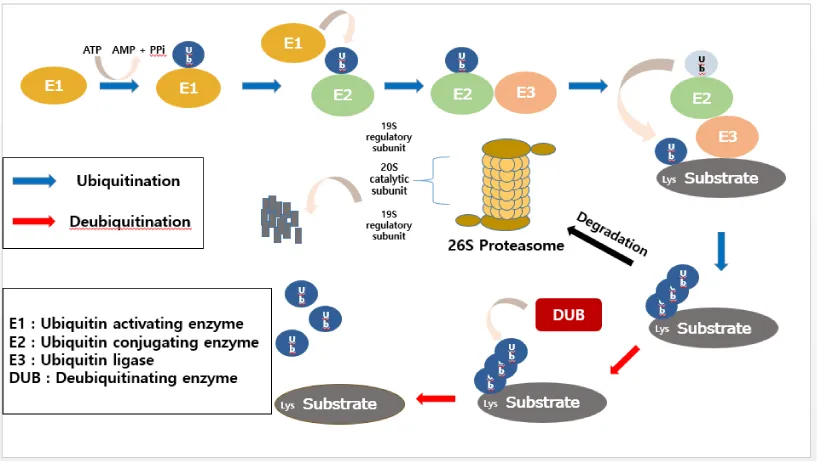
Ubiquitination is one of versatile post-translational modifications (PTMs), and it controls almost every aspect of cellular homeostatic processes in eukaryotic cells [1]. Therefore, it is pivotal to regulate the proper ubiquitin signaling for homeostatic maintenance [2]. In general, proteins degraded by the proteasome system are initially conjugated to the ubiquitin (Ub), a small polypeptide of 76 amino acids, which serves as a signal [3,4]. In a sequential events regulated by a cascade of ubiquitin-conjugating enzymes, ubiquitin is conjugated to specific substrates. The first class of enzymes involved in this cascade reaction is called the ubiquitin activating enzymes (E1). This E1-Ub complex then undergoes a reaction with one of ubiquitin carrier proteins (E2), in which the bound ubiquitin is transferred from E1 to E2. E2, in conjunction with an ubiquitin-ligase enzyme (E3), mediates the final transfer of ubiquitin to the target protein [5,6]. Covalent attachment of ubiquitin to target proteins induces their degradation by the 26S proteasome [7].
It has been identified that there are seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) in the peptide sequence of ubiquitin, and ubiquitin chains targeted for substrate proteins are formed on these residues serving as acceptors for another ubiquitin [8]. Different types of polyubiquitin chains generate various outcomes of the target protein [9]. When the K48-linked polyubiquitin chain, the most abundant linkage type, is established on target proteins, the 26S proteasome recognizes and degrades the polyubiquitinated protein [10]. In contrast, K63-linked polyubiquitination modifies diverse protein functions including DNA repair and endocytosis but does not lead to proteasomal degradation [11,12]. However, it is still not fully understood how different types of polyubiquitin chains are decoded in a cellular system yet.
Deubiquitinating enzymes (DUBs) play a role in removing ubiquitin molecules from substrates and have diverse roles in cellular processes [13]. It has been suggested that approximately 100 DUBs are encoded in the human genome [2], and they are classified into distinct subfamilies depending upon the organization of the catalytic domain. In general, they have been grouped into two main classes: cysteine protease and metalloprotease [14]. The cysteine protease class contains ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), ovarian tumor protease (OTU), Machado-Joseph disease protease (MJD), permutated papain fold peptidases of dsDNA viruses and eukaryote (PPPDE), and motif interaction with Ub-containing Novel DUB (MINDY1) family. The metalloprotease class is represented by the Jab1/Pab1/MPN metalloenzyme motif protease (JAMM) family [15-17]. Dysregulation of DUBs is becoming evident that they can be causative in a number of human diseases including cancer, and immune and neurodegenerative diseases [18]. Therefore, DUBs have been good targets to develop therapeutic drugs especially for diseases like cancer and immune diseases.
It has been demonstrated that cytokine-inducible DUBs, DUB-1, DUB-2, DUB-2A, DUB-1A, and DUB-3 (=USP17), are involved in the regulation of cellular proliferation and apoptosis [19-24]. These cytokine-inducible DUBs are relatively small enzymes of approximately 500 ~ 550 amino acids, sharing amino acid similarity not only in the Cys, Asp, and His domains for catalytic activity of the enzyme, but also their primary amino acid sequences [21,22]. It was demonstrated that DUB-1 is induced by cytokines IL-3, IL-5 and GM-CSF in B-lymphocytes, and it was found that DUB-1 is involved in cell growth arrest in the G1 phase of the cell cycle [19]. Interestingly, it was demonstrated that DUB-2 is induced by IL-2 only in T-lymphocytes [20]. And Dub-2 knock-out mice showed embryonic lethality and growth inhibition [25]. In addition, DUB-3 identified in human chorionic villi is induced by IL-4 and IL-6 and is involved in the regulation of cell growth and survival [23-25]. However, most of their substrates for these cytokine-inducible DUBs have not been found yet and their deubiquitinating enzyme activity has been mostly tested only in vitro. Since the enzymatic activity of these DUBs has been tested in vitro with the long period of induction to cleave the ubiquitin, it is not possible to know how fast the enzymatic activisty appears.
Since DUB-1, DUB-2, DUB-2A, DUB-1A and DUB-3, belonging to the subfamily of cytokine-inducible DUB, are involved in the regulation of cell proliferation and apoptosis as a part of interleukin-associated signaling pathways in lymphocytes [19-24], they are plausible targets from the therapeutic point of view in the context of homological oncology.
Conclusion
Spatial and temporal elimination of misfolded and damaged proteins is required to prevent the accumulation of toxic proteins, which can lead to diverse diseases including cancer and immune diseases [7]. However, it is not clear how a cell determines whether proteins targets should be degraded by the UPS or not. Given the significance of ubiquitin signaling in human diseases, important directions for future studies will include the analysis of underlying mechanisms that regulate the UPS for developing therapeutic implications for cancer and other immune diseases.
Even though cytokine-inducible DUBs are known to be involved in the regulation of cell proliferation and apoptosis, their DUB inhibitors have not been identified or developed for therapeutic purposes yet. Therefore, important directions for future studies will include identification of their substrates and underlying molecular mechanisms, and delineation of their molecular structures to provide us a novel way of developing therapeutic drugs especially for disease like cancer and immune diseases.
I would like to thank members of Baek laboratory for their critical comments on the manuscript. I also thank Jin-Tae Choi for making figure 1. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (2016R1A2B4008635).
- Karve TM, Cheema AK (2011) Small changes huge impact: the role of protein post-translational modifications in cellular homeostasis and disease. J Amino Acids 207691.Link : https://goo.gl/1TWHxy
- Kwon SK, Saindane M, Baek KH (2017) p53 stability is regulated by diverse deubiquitinating enzymes. Biochim Biophys Acta Rev Cancer. 1868(2) 404-411. Link : https://goo.gl/epWGvE
- Kwon YT, Ciechanover A (2017) The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci 42: 873-886. Link : https://goo.gl/ss2Gka
- Harper JW, Ordureau A, Heo JM (2018) Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 19: 93-108. Link : https://goo.gl/r19yZq
- Baek KH (2003) Conjugation and deconjugation of ubiquitin regulating the destiny of0020 proteins. Exp. Mol. Med. 35: 1-7. Link : https://goo.gl/Z13Hbw
- Wilkinson KD (1997) Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J 11: 1245-1256. Link : https://goo.gl/sdxzRq
- Leestemaker Y, Ovaa H (2017) Tools to investigate the ubiquitin proteasome system. Drug Discov Today Technol 26: 25-31. Link : https://goo.gl/iynWtA
- Woelk T, Sigismund S, Penengo L, Polo S (2007) The ubiquitination code: a signalling problem, Cell Div 2: 11.Linl : https://goo.gl/evaXx4
- Grice GL, Nathan JA. (2016) The recognition of ubiquitinated proteins by the proteasome. Cell Mol Life Sci 73: 3497-3506.Link : https://goo.gl/eULqJd
- Powell SR. (2006) The cardiac 26S proteasome: regulating the regulator Circ Rec 99: 342-245.Link : https://goo.gl/5SVVQ7
- Spence J, Sadis S, Haas AL, Finley D (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15: 1265-1273.Link : https://goo.gl/TY1xsT
- Wang G, Gao Y, Li L, Jin G, Cai Z, et al. (2012) K63-linked ubiquitination in kinase activation and cancer. Front Oncol 2: 5.Link : https://goo.gl/kPfGZM
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363-397. Link : https://goo.gl/33DLBu
- Hanpude P, Bhattacharya S, Dey AK, Maiti TK (2015) Deubiquitinating enzymes in cellular signaling and disease regulation. IUBMB Life. 67: 544-555.Link : https://goo.gl/bNQqan
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, et all. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell. 123: 773-786.Link : https://goo.gl/L5YqQa
- Iyer LM, Koonin EV, Aravind L (2004) Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 3: 1440-1450.Link : https://goo.gl/17nzF1
- Abdul RSA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, et all. (2016) MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 63: 146-155.Link : https://goo.gl/Rv4NQK
- King RW, Finley D (2014) Sculpting the proteome with small molecules. Nat Chem Biol. 10: 870-874.Link : https://goo.gl/YxszUp
- Zhu Y, Carroll M, Papa FR, Hochstrasser M, D’Andrea AD et all. (1996) DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc Natl Acad Sci U S A. 93: 3275-3279.Link : https://goo.gl/Zgsdfd
- Zhu Y, Lambert K, Corless C, Copeland NG, Gilbert DJ, et all. (1997) DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J Biol Chem. 372: 51-57.https://goo.gl/5fNd4N
- Baek KH, Mondoux MA, Jaster R, Fire-Levin E, D’Andrea AD et all. (2001) DUB-2A, a new member of the DUB subfamily of hematopoietic deubiquitinating enzymes. Blood. 98: 636-642. Link : https://goo.gl/hJBryU
- Baek KH, Kim MS, Kim YS, Shin JM, Choi HK et all. (2004) DUB-1A, a novel deubiquitinating enzyme subfamily member, is polyubiquitinated and cytokine-inducible in B-lymphocytes. J Biol Chem. 279: 2368-2376.Link : https://goo.gl/bm6gMn
- Burrows JF, McGrattan MJ, Rascle A, Humbert M, Baek KH et all. (2004) DUB-3, a cytokine-inducible deubiquitinating enzyme that blocks proliferation. J Biol Chem. 279: 13993-14000.Link : https://goo.gl/E8yRrB
- Shin JM, Yoo KJ, Kim MS, Kim D, Baek KH et all. (2006) Hyaluronan-and RNA-binding deubiquitinating enzymes of USP17 family members associated with cell viability. BMC Genomics. 7: 292.Link : https://goo.gl/k3xya5
- Baek KH, Lee H, Yang S, Lim SB, Lee W et all. (2012) Embryonic demise caused by targeted disruption of a cysteine protease Dub-2. 7: 044223.Link : https://goo.gl/hQaSZF
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
