International Journal of Immunotherapy and Cancer Research
A Study of Glucosylceramide Synthase and Glycolytic Pathway Enzymes in Patients with Hypereosinophilia
Lisa Redmond, Paul Kennedy, Colm Buckley and Philip T Murphy*
Cite this as
Redmond L, Kennedy P, Buckley C, Murphy PT (2017) A Study of Glucosylceramide Synthase and Glycolytic Pathway Enzymes in Patients with Hypereosinophilia. Int J Immunother Cancer Res 3(1): 015-018. DOI: 10.17352/2455-8591.000014Glucosylceramide synthase is the rate limiting enzyme in ceramide glycosylation and shifts the balance between ceramide and glycosphingolipids towards proliferation and survival of cancer cells. Increased glucose availability and glycolytic metabolism is preferentially used by cancer cells and has been linked to glycosphingolipid formation. We measured gene expression levels of glucosylceramide synthase and of glycolytic pathway enzymes, glucose transporter 1 and hexokinase II, as well as protein expression of glucosylceramide synthase, on trephine biopsy samples from 13 patients with hypereosinophilia, We found no significant differences in expression levels of any of the enzymes in patients with secondary causes of hypereosinophilia compared to those with either suspected or proven clonal hypereosinophilia.
Introduction
Eosinophilia, defined as a peripheral blood eosinophil count > 0.5 x 109/L, is frequently encountered in clinical practice. Reactive eosinophilia is a secondary cytokine driven phenomenon, due to a wide range of inflammatory, infectious, allergic and neoplastic conditions [1]. Less commonly, when there is an underlying stem cell, myeloid or eosinophilic neoplasm, eosinophilic cells are considered clonal [1,2].
According to recent consensus proposals, hypereosinophilic syndrome (HES) is defined as (i) hypereosinophilia (HE) (eosinophil count >1.5 x 109/L), documented on at least 2 occasions, or marked tissue eosinophilia, (ii) clinical manifestations attributable to the eosinophilia, and (iii) the absence of an alternative explanation for the observed organ damage [3]. HES can be divided into primary (neoplastic) (HESN), where eosinophils are considered neoplastic (clonal), secondary (reactive) (HESR), where eosinophilia is non-clonal and cytokine driven by an underlying condition/disease and idiopathic HES, with no evidence of an underlying reactive or neoplastic condition [3]. In addition, for patients with unexplained persistent asymptomatic HE, the term hypereosinophilia of unknown significance (HEUS) has been coined. Such patients have no evidence of an underlying hereditary, reactive or neoplastic condition but also no evidence of eosinophil-related organ damage and their prognosis is uncertain [3].
Although some patients with HESN are relatively easy to diagnose, such as patients with chronic eosinophilic leukemia (CEL) with the FIP1L1-PDGFRA fusion gene and exquisite sensitivity to imatinib [4], clonality may be difficult to detect in many other cases of suspected HESN, and some cases of clonal eosinophilia may thus be assigned to the category of idiopathic HES or HEUS.
Ceramide and glycosphingolipids (GSLs) are important biological molecules in cellular processes of cancer progression due to their effects on various aspects of cell function such as apoptosis (programmed cell death), proliferation and migration [5]. The balance between ceramide and GSLs can induce cancer cells to proliferate or to undergo apoptosis [6]. Glucosylceramide synthase (GCS) (also known as UDP-glucose ceramide glucosyl transferase (UGCG)) is the rate limiting enzyme in ceramide glycosylation by shifting reactions to generate metabolites in favour of cancer [7]. Following chemotherapy or radiotherapy, an increase in ceramide leads to proliferation arrest and apoptosis of the cancer cells [8]. However, enhanced ceramide glycosylation, by promptly converting pro apoptotic ceramide into pro survival glucosylceramide, inhibits these processes and is one recognised mechanism of drug resistance in cancer [8]. GCS has been shown to be upregulated in many cancer cells [8]. In chronic lymphocytic leukemia (CLL), the most common leukemia, B cell receptor triggers apoptosis resistance of primary CLL cells by upregulating GCS and thus, reducing the ratio of ceramide to glucosylceramide [9]. Unlike patients with reactive eosinophilia, cultured eosinophils from patients with HES have delayed spontaneous apoptosis and relative resistance towards ceramide- but not CD95-mediated death [10].
Cancer cells preferentially utilise increased glucose availability and glycolytic metabolism to produce ATP, with Glucose transporter 1 (GLUT1) and Hexokinase II (HK2) frequently overexpressed to take up elevated amounts of glucose [11]. A definite link has now been shown to exist between glucose availability and GSL formation [12]. In a leukemia cell line with elevated glucosylceramide, treatment with inhibitors of glycolysis or the pentose phosphate pathway significantly decreased glucosylceramide [12].
In this study, we compared gene and protein expression levels of GCS in patients with HESN, idiopathic HES and HEus to those of patients with secondary HE to investigate the possibility that patients with proven or suspected clonal eosinophilia, in contrast to those with secondary eosinophilia, have increased levels of GCS, resulting in a reduced ceramide: glucosylceramide ratio with consequent resistance to apoptosis and increased proliferation. As well as being of diagnostic use, the demonstration of increased GCS in patients with clonal eosinophilia might also be of therapeutic benefit as the combination of ceramide with a GCS inhibitor might be an effective therapeutic strategy [13]. We also measured gene expression levels of GLUT1 and HK2 as measures of increased glucose uptake.
Materials and Methods
After study approval by Beaumont Hospital Ethics Committee, 13 consecutive patients with eosinophil counts > 1.5 x 109/L gave written informed consent for this study.
All patients had the following tests performed at diagnosis: peripheral blood samples for complete blood count, serum immunoglobulins, including total serum IgE, serology for helminths, autoimmune screen, including antineutrophil cytoplasmic antibodies (ANCA), serum tryptase, T cell immunophenotype and T cell receptor gene rearrangement studies and JAK2V617F mutation; bone marrow aspirate, trephine biopsy and bone marrow cytogenetics, including FISH probe to detect interstitial deletion on chromosome 4q12, which results in the formation of the FIP1L1-PDGFRa fusion gene; CT scan of chest, abdomen and pelvis.
Total RNA extraction from archive formalin-fixed paraffin-embedded (FFPE) diagnostic bone marrow trephine tissue blocks was performed using the miRNeasy FFPE kit from Qiagen according to the manufacturers’ instructions. The concentration and quality was evaluated by a Nanodrop. cDNA was synthesized from 2µg of total RNA using the Applied Biosystems high capacity reverse transcription kit which was performed on a G-storm thermocycler. Primer and probe for the GCS (12), GLUT1 and HK2 [14], were synthesised by Eurogentec. Standards were synthesized by Euro fin.
For real time PCR, 4 μl cDNA was mixed with 12.5 µl Taqman fast universal master mix (Applied Biosystems), 400nM specific oligonucleotide primers (Eurogentec) and 5.5 µl dH2O (Sigma) to a final volume of 25 µl. An internal housekeeping gene control, GAPDH was used to normalize differences in RNA isolation, RNA degradation, and the efficiencies of the RT (Applied Biosystems). PCR reaction was carried out as follows: 95oC for 30s; denaturation 95oC 5s; annealing 60oC for 5s; 40 cycles in total. Primers and probes were validated for efficiency prior to use in qPCR. The sequences of the primers and probes are shown in table 1.
Immunohistochemical staining for GCS protein expression was performed on slides from the archive FFPE diagnostic bone marrow trephine tissue blocks. Staining was performed using Anti-GCS antibody (Sigma-Aldrich) on the benchmark ultra-platform using optiview detection kit with amplification at a dilution of 1:100 with 90 minute heat mediated antigen retrieval.
Statistics
The Mann-Whitney U test was used to compare differences between two independent groups. Pearson correlation coefficient was used as a measure of linear dependence between two variables. P values < 0.05 were considered statistically significant.
Results
Based on the results of investigations listed in the Materials and Methods section, the following diagnoses were made: CEL with FIP1L1-PDGFRA fusion gene (n=1); idiopathic HES (n=3); HEUS (n=2); secondary eosinophilia (n=7) (Table 2). Three of the cases of secondary eosinophilia had an underlying diagnosis of T cell lymphoma but none of the cases had bone marrow involvement by lymphoma.
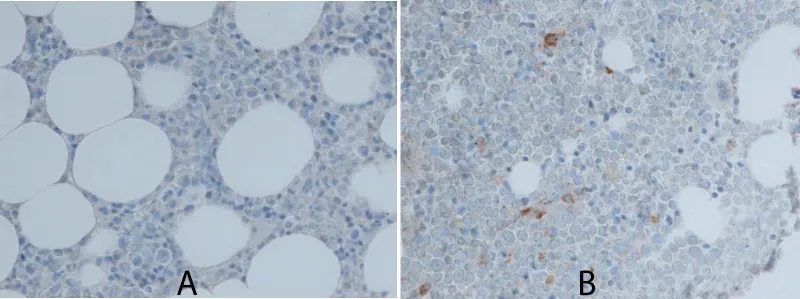
No significant differences in gene expression levels of GCS, HK2 and GLUT1 or in protein expression of GCS were found between patients with secondary eosinophilia and the group of patients with CEL, idiopathic HES and HEUS (Table 2). In addition, there were no statistically significant correlations between peripheral blood eosinophil count at diagnosis and gene expression levels of either GCS, HK2 or GLUT1. Images of patient trephine biopsy slides, positive and negative for GCS protein expression, are shown in figure 1.
Discussion
In many cases of suspected clonal eosinophilia, conventional methods to demonstrate clonality such as conventional cytogenetics and FISH fail to show a molecular lesion [15]. This has led others to seek alternative means to try to differentiate clonal from reactive eosinophilia. Cilloni, et al., have reported that WT1 transcript amounts can discriminate between HES/CEL and reactive eosinophilia in a study of 312 patients with hypereosinophilia [16], whilst Andersen, et al., have suggested that DNA methylation signature may help to distinguish clonal and suspected clonal eosinophilia from reactive eosinophilia [17].
In our study, we hypothesised that cases of clonal hypereosinophilia would have evidence of a reduced ceramide: glucosylceramide ratio and of increased glucose uptake. The finding of increased GCS expression might have proved useful in helping to establish a diagnosis in patients with suspected clonal eosinophilia and attempts at targeting of ceramide glycosylation might have emerged as a potential therapeutic strategy. However, our results failed to show any difference between mRNA expression levels of GCS, hexokinase and GLUT 1 or in GCS protein expression in cases of suspected or proven clonal eosinophilia compared to cases of reactive eosinophilia. Drawbacks to our study include the very small number of cases studied and the undoubted heterogenous nature of cases that fall into the category of idiopathic HES and HEUS, with a substantial number of these cases probably not due to clonal eosinophilia. In addition, our study retrospectively measured parameters of the GCS system on trephine biopsies of patients with eosinophilia, rather than on purified eosinophils obtained from peripheral blood or bone marrow aspirate, making it less likely to detect any possible difference between clonal and non-clonal eosinophils due to contamination by other bone marrow hematopoietic and stromal cells.
Despite these negative findings, the report of delayed spontaneous apoptosis and resistance to ceramide in cultured eosinophils from patients with HES [10], suggests that the ceramide pathway in certain subsets of clonal HES might still be of potential diagnostic and therapeutic importance and that further such studies of the ceramide/GSL system in purified eosinophils from larger numbers of patients with both clonal and reactive eosinophilia should be considered.
- Tefferi A, Patnaik MM, Pardanani A (2006) Eosinophilia: secondary, clonal and idiopathic. Br J Haemat 133: 468-492. Link: https://goo.gl/6frWC3
- Bain BJ, Fletcher SH (2007) Chronic eosinophilic leukemias and the myeloproliferative variant of the hypereosinophilic syndrome. Immunol Allergy Clin North Am 27: 377-388. Link: https://goo.gl/x2nO0D
- Valent P, Klion AD, Horny HP, Roufosse F, Gotlib J, et al. (2012) Contemporary consensus proposal on criteria and classification of eosinophilic disorders and related syndromes. J Allergy Clin Immunol 130: 607-612. Link: https://goo.gl/JST22V
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, et al. (2003) A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypersosinophilic syndrome. N Engl J Med 348: 1201-1214. Link: https://goo.gl/ZvnHz7
- Liu YY, Hill RA, Li YT (2013) Ceramide glycosylation catalyzed by glucosylceramide synthase and cancer drug resistance. Adv Cancer Res 117: 59-89. Link: https://goo.gl/SlMI5c
- Ogretmen B, Hannun YA (2004) Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer 4: 604-616. Link: https://goo.gl/W2h84G
- Larsen PJ, Tennagels N (2014) On ceramides, other sphingolipids and impaired glucose homeostasis. Mol Metab 28: 252-260. Link: https://goo.gl/e7rTPA
- Chapman JV, Gouazé-Andersson V, Messner MC, Flowers M, Karimi R, et al. (2010) Metabolism of short-chain ceramide by human cancer cells – implications for therapeutic approaches. Biochem Pharmacol 80: 308-315. Link: https://goo.gl/qtCDYY
- Schwamb J, Feldhaus V, Baumann M, Patz M, Brodesser S, et al. (2012) B-cell receptor triggers drug sensitivity of primary CLL cells by controlling glucosylation of ceramides. Blood 120: 3978-3985. Link: https://goo.gl/mR9LY5
- Vassina EM, Yousefi S, Simon D, Zwicky C, Conus S, et al. (2006) cIAP-2 and survivin contribute to cytokine-mediated delayed eosinophil apoptosis. Eur J Immunol 36: 1975-1984. Link: https://goo.gl/NcWoAC
- Aloj L, Caracó C, Jagoda E, Eckelman WC, Neumann RD (1999) GLUT1 and hexokinase expression: relationship with 2-fluoro-2-deoxy-D-glucose uptake in A431 and T47D cells in culture. Cancer Res 59: 4709–4714. Link: https://goo.gl/gBqino
- Stathem M, Marimuthu S, O’Neal J, Rathmell JC, Chesney JA, et al. (2015) Glucose availability and glycolytic metabolism dictate glycosphingolipid levels. J Cell Biochem 116: 67-80. Link: https://goo.gl/nDi3HC
- Watters RJ, Fox TE, Tan S-F, Shanmugavelandy S, Choby JE, et al. (2013) Targeting glucosylceramide synthase synergizes with C6-ceramide nanoliposomes to induce apoptosis in NK leukemia. Leuk Lymphoma 54: 1288-1296. Link: https://goo.gl/4KjIUj
- Binderup T, Knigge UP, Federspiel B, Sommer P, Hasselby JP, et al. (2013) Gene expression of glucose transporter 1 (GLUT1), hexokinase 1 and hexokinase 2 in gastroenteropancreatic neuroendocrine tumors: correlation with F-18-fluorodeoxyglucose positron emission tomography and cellular proliferation. Diagnostics 3: 372-384. Link: https://goo.gl/O4evoq
- Bain BJ (2004) Relationship between idiopathic hypereosinophilic syndrome, eosinophilic leukemia and systemic mastocytosis. Am J Hematol 77: 82-85. Link: https://goo.gl/MzqShR
- Cilloni D, Messa F, Martinelli G, Gottardi E, Arruga F, et al. (2007) WT1 transcript amount discriminates secondary or reactive eosinophilia from idiopathic hypereosinophilic syndrome or chronic eosinophilic leukemia. Leukemia 21: 1442-1450. Link: https://goo.gl/c27210
- Andersen CL, Nielsen HM, Kristensen LS, Søgaard A, Vikeså J, et al. (2015) Whole-exome sequencing and genome-wide methylation analyses identify novel disease associated mutations and methylation patterns in idiopathic hypereosinophilic syndrome. Oncotarget 6: 40588-40597. Link: https://goo.gl/AkBruv
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
